MARCO Polo: Finding Crystals
Published September 30, 2020 This content is archived.
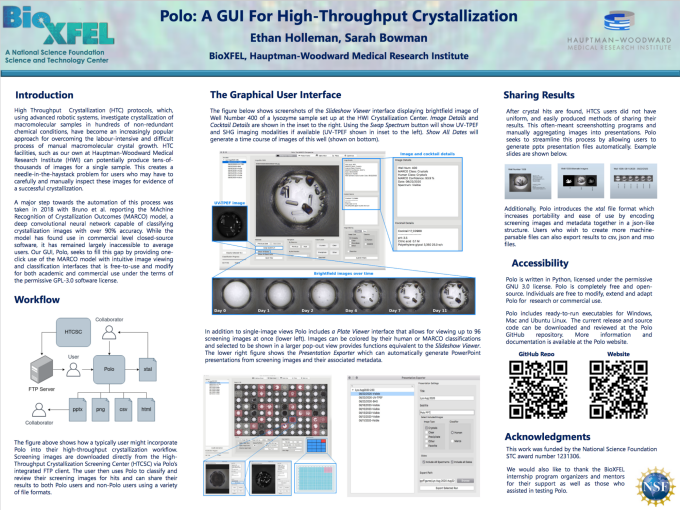
Summer intern Ethan Hollerman won a Student Poster Prize at the 2020 LCLS/SSRL Users’ Meeting for his work on implementing the MARCO algorithm into a new GUI! MARCO POLO is a multi-platform open-source Python-based graphical user interface that has been developed to provide access to MARCO automated classification and data management tools for biomolecular crystallization screening. MARCO is an academic and industry collaboration using image data from the Crystallization Center, Bristol-Myers Squibb, GalaxoSmithKline, Merck, and the Collaborative Crystallization Centre in Australia. It involved image analysis experts at Google Brain, The University at Buffalo, Duke University, and the University of York in the UK, with industry input from Formulatrix. MARCO is being actively used in industrial settings, in Formulatrix instruments, and soon for users of the Crystallization Center. The GUI is undergoing beta testing, a paper is forthcoming, and the GUI will be released shortly.